The longer I live, the wronger I get to be
On the gift of an emerging field’s incessant pace of self-correction
At a societal level, cursing an enemy to "live in interesting times" can feel just as cruel as intended. But there are probably few things more bracing than living through "interesting times" in your field of choice. Imagine if we could transport some of the late luminaries of the field of human population genetics from their relatively uneventful eras to our fervent times instead. Think if minds of that caliber could dig into the fine-grained detail of everything their statistically brilliant, but cruelly data-deprived models foretold. Titans like L.L. Cavalli-Sforza, W. D. Hamilton, Sewall Wright and R.A. Fisher concluded their contributions to the field without ever beholding the avalanches of undifferentiated riches we now ceaselessly sift through.
Of course, among the downsides of interesting times is that from right at the heart of the maelstrom, you get constantly pelted with a whole lot of mess and confusion. If your polestar is enduring personal achievement/contribution or optimizing your ratio of good bets to bad, or if you only like your debates long since settled, interesting times might truly be a curse. If, on the other hand, your bent is simply to drink down as much data and insight as one lifetime will allow, you could do worse than interesting times.
These past two decades in human population genetics have been dizzyingly interesting to me, rife with plot twists, unforeseen reversals and extreme course corrections, the way forward strewn with the corpses of mercilessly dethroned theories. I can only look back over this young century in my field with awe and an enormous dose of humility. Because if I am anything, it is a gadfly, a wide-eyed super fan, the guy who has a take on everything, inhales every supplement, scrutinizes every method and is always happy to publicly bet on a horse. Which entails a lot of being wrong in public. And so what? It comes with the territory. In the end, I am truly just here for the raw data, following along with fascination wherever it leads us. I have been wrong about big questions in this field so many times already in scarcely a quarter century of post-collegiate life. And what better option do I have than to stand corrected every time, as our understanding of what it means to be human proceeds forward in unpredictable lurches, painstaking rewrites and spasms of brilliant insight.
Below are some key cases in the field where I can look back and say I stand corrected, sometimes even just a few productive years later. Where there is one, I include a relevant paper or podcast conversation. Here's to the interesting times rolling unpredictably on!
From simple “Out of Africa” to it’s complicated, but still a lot of “Out of Africa”
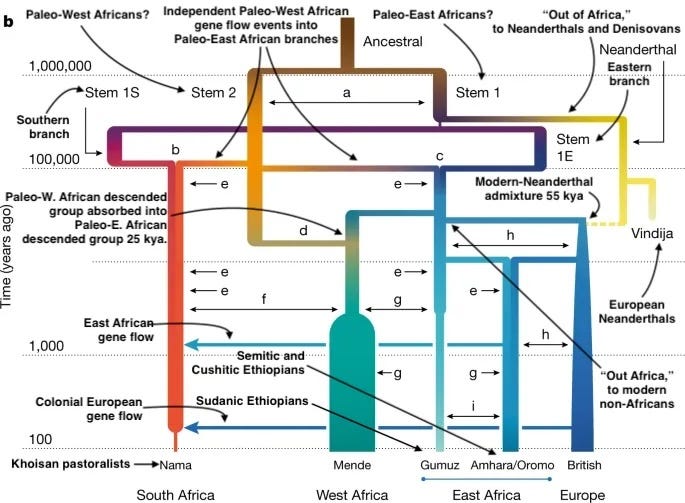
I no longer believe that modern humans emerged in a single contingent speciation event 50-200,000 years ago in Africa. Instead, what it means to be or to have been human is the outcome of countless small changes accumulating gradually over the last million years, part of a broader shift that began with our common ancestors with Neanderthals and Denisovans and encompassed all three of our parallel lineages. While it is true that some periods saw shifts in the rate of cultural evolution (like the “cognitive great leap forward” about 40,000 years ago, memorably accompanied as it was by symbolic art), what we perceive to be a stark revolutionary break was actually probably just part of a continuous exponential change, as human cultures mutated and evolved ever faster. Our lineage’s origin had a very long fuse back to the origins of Homo, even if the arrival of our modern lineage looms up in our perceptual frame as a leap forward. Consider that just like our own ancestral paleo-African lineage, Neanderthals were also evolving larger and larger brains over the last few hundred thousand years up until their extinction. A general force toward greater cognitive ability and cultural complexity was sweeping across our whole competing lineage of also-rans, not just differentiating those lines of humans who survived and gave rise to us. We weren’t special, someone with the secret sauce was inevitably going to win out... and since we all had it, perhaps we need to start thinking of ourselves more as lucky than special. Related post: Current status: complicated.
No one big “IQ gene,” just lots and lots of minor ones
At the turn of this century, the media and scientists were still holding their breath for “IQ genes” to be pinpointed. In 1998 The New York Times published First Gene to Be Linked With High Intelligence Is Reported Found. Studies like this were cited to great fanfare in Matt Ridley’s otherwise remarkably misstep-proof 1999 classic Genome: The Autobiography of a Species in 23 Chapters. Looking back in the 2020’s, it is clear these were never anything but statistical flukes. No known genes result in massive boosts to IQ; studies purporting to show this invariably turn out to have small sample sizes (this stands in contrast to massive mutations that can cause mental disability). For example, the 1998 study cited above used a small sample size of individuals with high IQs, just N = 50. Now we know intelligence resembles height dynamics, except ratcheted up a notch: the effects on the characteristic are distributed over thousands of positions across the genome, with mostly small, almost imperceptible, effects. IQ remains well approximated by R. A. Fisher’s 1918 infinitesimal model; it is proving to be a quantitative trait better understood through statistical theories than with investigation via empirical molecular analysis. Related podcast: Alex S. Young and James J. Lee: quantitative genetics in 2023.
European Jews are not just converts, they are a demographic synthesis
Perhaps a slightly obscure thing to be wrong about, but in the early 2000’s I could still read in older genetics textbooks that European Jews were mostly descended from Europeans. Obviously this is not quite the case, Ashkenazi Jews have a lot of Middle Eastern ancestry. With hindsight, I feel like I should have seen this coming, especially given that the Cohen Modal Haplotype thesis, where men with the surname Cohen share the same ancestor dating to 3,000 years ago, was already out in the late 1990’s, indicating deep Middle Eastern roots for the Ashenazim. Related post: More than kin, less than kind: Jews and Palestinians as Canaanite cousins.
From one expansion to many (and all the admixtures)
In 2004, medical doctor and geneticist Stephen Oppenheimer published The Real Eve: Modern Man’s Journey Out of Africa. Oppenheimer presented a narrative based on mitochondrial DNA, human maternal lineages, where our lineage expanded out of Africa during the last Ice Age 80,000 years ago, and settled various regions of the world before diversifying in each. In Oppenheimer’s model, aside from the post-Columbian migrations, most of the world’s population structure and variation was already established during the Pleistocene, which ended 11,700 years ago. When I read The Real Eve I accepted this model as a given. But paleogenetics has subsequently brought many new findings that have forced a reconsideration. Massive migration and admixture over the last 11,700 years are undeniable, with events like the Bantu expansion barely predating the New World’s post-Columbian migrations. Europe, where we have good ancient DNA samples across the last 45,000 years has seen at least a half dozen wholesale population turnovers since the first modern humans arrived. The narrative in The Real Eve turns out to have been merely prologue, not holy writ. Related paper: Toward a new history and geography of human genes informed by ancient DNA.
From the end of natural selection to its resurrection. Selection strikes back!
Humans are a large slow-breeding population. We live in a world where infant mortality has sharply crashed, and family size is mostly a matter of choice. In the year 2000, I trusted that natural selection was not a major force on human evolution in our time. First, since our small population meant we were drift-dominated, random changes should be dominant even without the conditions of modernity. Second, exactly what selection pressures applied in the late 20th century? These views remain rather common, but I no longer share them, because I think natural selection remains ubiquitous for our species. First, local adaptation seems to have occurred all across the world on relatively short time scales, from circadian rhythms in northern Eurasia (the last 100,000 years) to cholera adaptations in Bangladesh (the last 1,000 years) and malaria resistance in Madagascar (the last 1,000 years). The emergence of genomics as a field has resulted in several back and forths about the role of adaptation in human evolution, but overall I see good evidence for natural selection being critical to our immune and digestive systems no matter who you ask. We have the molecular evidence in the DNA sequences now in a way we did not twenty years ago. Additionally, we now know selection operates in different ways for different traits. Lactase persistence is targeted at a single gene and at least in Europe, Central Asia and South Asia is focused on a single mutational variant from a common ancestor. But other traits, like height, seem to have been under selection for much longer, and are distributed across many genes with diverse origins. In the case of quantitative traits like height, selection works on old variation rather than new mutations.
The second thing to note is that even today with low infant mortality, we still observe high miscarriage rates. The mutations vying for admittance into the next generation still have to make it through a major selective sieve that we don’t see or measure because it’s all in utero. Finally, the reproductive variance in cultures we see also has a long evolutionary impact. I suspect that after 200 years of selection (a good eight generations) the Amish today (whose fertility rates are on the order of 6-7 children per woman) are somewhat different from the Amish of 1800. Over the last few centuries, the number of Amish leaving their communities has been declining every generation, perhaps due to selection for particular personality types that are disinclined to defect from such a closed-off and conformist community.
Just find the gene and fix the disease…if only it were that simple
During the early years of the Human Genome Project, something called the common-disease common-variant hypothesis was widely promoted as our path to discover most of the causal genes for most diseases (it was to a great extent the funding justification for early human genomics in the 21st century). This model held the promise of a single test yielding a wide panoply of risks and probabilities for everyone. In 2024 we still don’t have good tests for many diseases, and risks for many conditions like type 2 diabetes or autism are the outcome of many genes across many loci. The problems here are not simple. In some cases there are so many variants that it is not necessarily easy to define all the causal effects and create a simple and powerful statistic with any individually predictive power. In other cases, the variants might be rare within the population, and therefore not in the catalog of disease-causing variants. This doesn’t mean the problem is intractable; it’s a matter of sequencing many more humans and collecting medical data on the sequenced individuals. But this looks to be at least a two-generation rather than a single generation project, as initially projected. Arguably we have all the technology in place, from cheap sequencing to powerful computational techniques, but a focused international project may need to coalesce to push research across the finish line.
We aren’t Neanderthals…to we kind of are Neanderthals (a bit)
If I’d been forced to guess in 2004, I would have said humans probably didn’t have ancestry from Neanderthals, and I certainly wouldn’t have guessed that something like Denisovans existed. Part of this was the era’s regnant Out-of-Africa anthropocentrism. But some was a lack of appreciation of the possibility of hybridization across very distinct lineages. In 2005, I read Jerry Coyne and H. Allen Orr’s Speciation, and began to rethink my priors in this area. Further conversations with Greg Cochran and John Hawks also left me more open to the idea of admixture between our own lineage and Neanderthals, so that by 2010 when the first Neanderthal genome confirmed this phenomenon, I was not shocked, though I still might have been as excited as anyone that Svanto Paabo and colleagues had actually proven it with ancient DNA. Related post: Here be humans.
Genetics as an abstract science…to genetics as commodity
This one is simple: even in 2004 I did not anticipate that companies like 23andme, Family Tree DNA and Ancestry would take consumer genetics so far, with tens of millions of customers. When Spencer Wells organized the Genographic Project in 2005, I had no sense a new age of genomics was dawning, with research driven by consumer enthusiasm and participation. The idea of people paying for their own kits to drive research, and this phenomenon becoming so integrated into popular culture and resulting in millions of human samples was not even comprehensible as this century dawned. This is a whole new way of doing science. And clearly one of my favorite things to have been proven wrong about. Related comment: Consumer genomics will change your life, whether you get tested or not. So you want to test your DNA.
Gene interactions might explain a lot of evolution…but not on the species-level
About twenty years ago I was interested in epistasis. Not epigenetics, but epistasis, the phenomenon whereby the impact of variation at a gene of interest is conditional on the precise variation at other genes. A simple way to understand it is that the model where genetic variation has additive and independent effects (which, for example, works with intelligence and most quantitative traits) is a linear framework where each change accumulates independently and predictably. To calculate expected height or intelligence, just tote up the variants of interest. In contrast, in an epistatic system you can’t just assume that a given variant will have a given effect without knowing the “genetic background,” the variation across other genes that are interacting in a network.
To add to the confusion, molecular and evolutionary biologists often think of epistasis differently. Molecular geneticists are concerned with a sequence of processes across a pathway that link various genes, or perhaps direct interactions between the gene products at two loci. Evolutionary geneticists are more concerned with the overall fitness or phenotypic outcomes. It is the latter that particularly interested me, and led me to read Epistasis and the Evolutionary Process in 2005. Not to get too far into the weeds, but epistasis plays a large role in much of Sewall Wright’s later thinking on the ‘shifting balance’ of the adaptive landscape. Two decades on, I think statistical epistasis is not really that important or tractable for a lot of microevolutionary questions. This means that evolutionary change across “short” time scales like tens of thousands of years reflects adaptation within a species that is mostly additive and linear, and that’s what chiefly interests me now (though genetic interactions probably matter a lot for macroevolutionary between-species time scales). Related paper: Widespread signatures of natural selection across human complex traits and functional genomic categories.
From a surfeit of theory to a flood of data
This final one isn’t so much my own mind changing as the landscape of the field shifting in a way I still find amazing. Pausing to look back over these whirlwind decades, I think we can see that in short order a funny balance has shifted from one extreme to the inverse in human population genetics. For decades, ours was a theory-rich and comparatively data-poor field; these hyper-productive years have seen us churn through that accumulated backlog of theories waiting to be tested against fresh reams of data. Now, we have instead reached such a buildup of data that we are actually running short on theory to explain it. I never could have anticipated a situation where data generation and cursory analysis outpaced deep, slow-cooked theoretical understanding of how that data might snap into a bigger interpretative picture. Evolutionary population geneticist Matt Hahn stated nearly a decade ago in the mid 2010’s that in his particular subfield of molecular population genetics there was so much data that they could stop sequencing organisms, and it would still take decades to analyze the already pending results and integrate them into a theoretical framework. This is why AI-assisted science is probably going to become indispensable sooner rather than later, if we’re going to have any chance of increasing human productivity to match the rate of data generation.





I always joke to my students that everyone I taught about the settlement of the Americas at the start of my career could ask for their money back, because basically everything I said then has turned out to be wrong. It's been exciting along the way! But I hope my former students have either read the news about it or forgotten everything I said :)
It's really, really, such an exciting time for understanding the whole human story. So much of anthropology is a total mess, but that there is so much interesting stuff to be figured out gives me hope that it will simply have to turn itself around -- or be replaced as a discipline by scholars who will.
Small point. You mentioned that Cohanim are Jews with the surname Cohen. That is not true. Cohanim can have any surname. The status is passed down verbally from father to son. I am a Cohan, it has been verified by YDNA analysis. But my name is not Cohen. From what I understand Ashkenazi Jews did not adopt surnames until the late 18th century.