We were selected: tracing what humans were made for
Counting down 2024’s three biggest ancient DNA findings: number 3
The ancient-DNA era for the human species is not yet 15 years old. It kicked off with the 2010 paper Paleo-Eskimo genome (followed by blockbusters on Neanderthals and Denisovans). Today, remains from tens of thousands of ancient humans offer us decipherable DNA information, each contributing to fill in gaps in our understanding of prehistory.
No matter the brilliance and insight of its practitioners and theorists, human prehistory’s age of genetic inference, when we were limited to examining modern genomes to learn about ancient peoples, was like trying to comprehend the world’s oceans solely from the activity observable in its brightly lit uppermost layers, the photic zone that reaches down at most about 200 meters. Though some key insights date to the before times, the paleogenomic era after 2010 has been absolutely revolutionary and transformative. Name a superlative, and it probably isn’t strong enough.
This year, year 14 of our ancient-DNA golden age if you’re keeping count, was no exception to the relentless pace of revolution and transformative progress in the field. And yet, from my vantage, three topics charted particularly spectacular gains this year. As 2024 gives way to 2025, I have three short pieces for you sharing my picks for this year’s most exciting leaps forward in ancient DNA.
I have no doubt that my first pick, a September 2024 preprint will prove a landmark in the field. The culmination of years of work, it takes ancient DNA far beyond phylogeny, throwing open the gates to vast new landscapes of evolutionary dynamics. Humans are an expansive species that has spread across the planet, adapting to every locale, and this paper now turns our long history into one of evolution’s premier laboratories.
We were selected: tracing what humans were made for
Charles Darwin’s theory in On the Origin of Species was predicated on the power of natural selection to drive adaptation and diversification. Over 160 years later we understand selection in much greater detail than we did then. Agricultural scientists and farmers use selection to obtain desired outcomes, with the various attendant side effects (e.g., chickens so fat they are also sterile or sheep that produce a great deal more wool, but at much lower quality) well known by now. Meanwhile, in climate-controlled laboratories, for decades now geneticists have been manipulating lineages of fast-reproducing organisms like Drosophila melanogaster, the “fruit fly.” To understand myriad complex evolutionary dynamics, it is to drosophila that we have most often looked.
At least until this century. Genomics and ancient DNA are now finally allowing scientists to study humans like they would the fast-breeding fruit fly. Freed of the need for controlled breeding, laboratory experiments, fast generation time, or even patience, researchers now interrogate the whole human DNA sequence, comparing all three billion pairs across thousands of individuals. Novel statistical methods are being developed to meet the possibilities presented by the massive emerging surfeit of data, as researchers look for deviations and broader patterns, and try to smoke out the role of selection within the genome. To understand natural selection in the genome means tracing out new mutations’ arc of variation. Fruit fly generations are 10 days, offering the vantage of some 70 generations in the space of a 2-year research project. Going back to the advent of fruit fly research meanwhile would offer 4,234 generations. In human terms that’s like 1,800 years and 105,000 years respectively. With ancient DNA we can now examine selection in our species by reviewing the natural experiments of our past. It’s true, we don’t have controlled laboratory experiments, but we do have a species that has expanded across six continents, innovating and adopting countless different lifestyles along the way. And little by little, we accumulate more and more ancient samples of that species’ tell-all DNA.
Now, a September 2024 preprint, Pervasive findings of directional selection realize the promise of ancient DNA to elucidate human adaptation, marshals all the new techniques in human paleogenomics to tackle the question of how much and what kind of natural selection has shaped our species in our recent evolutionary past. The authors looked at 8,433 West Eurasians sampled across the past 14,000 years as well as 6,510 contemporary samples, focusing particularly on Europe because that continent offers so many samples across numerous time periods. Instead of detecting patterns in the present day, and testing how they fit hypothetical mathematical or computational models, the massive ancient DNA dataset allows them to detect changes over time, as new variants arise and frequencies fluctuate. Since the bulk of changes in gene frequencies owe to random genetic drift, the team specifically hunted for long-term shifts of variant frequency across populations that could not be explained simply by the populations’ own internal levels of relatedness. A steppe variant increasing in Europe because of steppe people’s mass arrival is not natural selection, but a change driven by the influx of new ancestry injecting new variants associated with new relatedness networks. Melanesians retaining the same skin tone as Africans despite being much closer genetically to Europeans is natural selection; (evidence from multiple methods makes clear that Melanesians and other darker-skinned Eurasians retain the ancestral state while Europeans, West Asians and East Asians have all grown paler in the last 10,000 years).
Even using stringent statistical filters, which produce a lower value than the true underlying phenomenon, the group found 347 independent loci of selection. Not unsurprisingly, 279 were in HLA regions. HLA stands for Human Leukocyte Antigen, and is involved in controlling variation in a major part of the adaptive immune system (if you get an organ transplant, the HLA match is crucial). Because large organisms are constantly co-evolving with their pathogens, the HLA region is very diverse and always evolving. If your test to detect natural selection doesn’t turn up copious results in the HLA region, your test is not very good.
But what about the other selective events besides HLA? Most of us know the ABO blood group system because of matching during donation (O is a universal donor, A can only donate to A and AB, B to B and AB, and AB solely to AB). Detecting whether you have the A or B antigen is simple, so the ABO system became one of the first major human population-genetic assays, popularized in the 1950’s. The fact that the letters refer to antigens belies their association with the immune system. But there are other implications as well, people who are A or B are at greater risk for heart disease, stroke, pancreatic cancer, malaria, COVID-19, blood clots and cognitive decline. So why do we have A and B, which are new mutations against the ancestral O background? Probably because of past plagues and endemic diseases. Blood group O is more susceptible to cholera and diarrheal diseases, for example. Who wouldn’t trade an upped risk for pancreatic cancer later in life to avoid dying of cholera early in life? This preprint shows that blood group B rose in frequency in the late Neolithic, starting 6,000 years ago, specifically at A’s expense. Though the precise mechanisms remain to be worked out, probably B is more protective against newer endemic diseases against which A had few defenses.
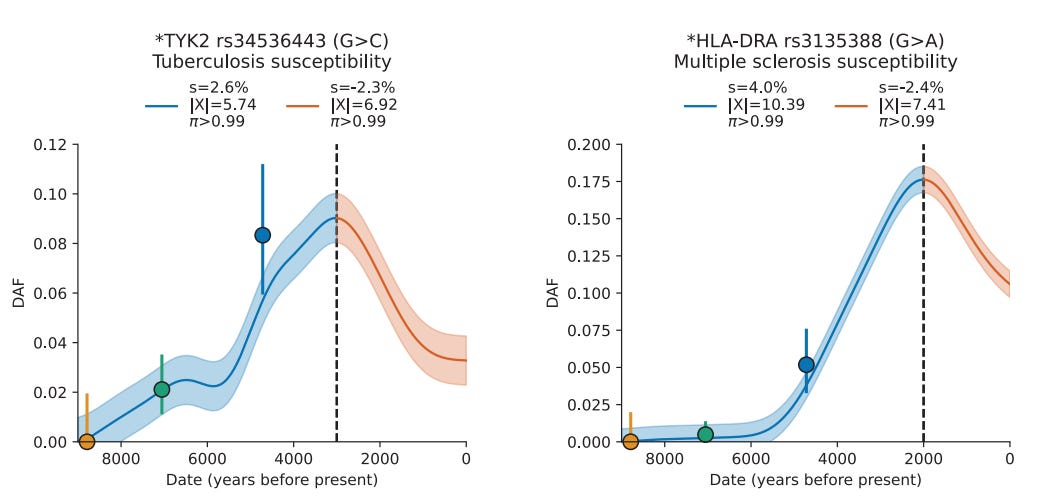
One of the major upsides of this large dataset spanning over 14,000 years is that the authors were able to detect reversals in natural selection too subtle and remote to infer solely from modern populations. Genetic variants implicated in relative susceptibility to tuberculosis, multiple sclerosis and Crohn’s disease shot up during the late Neolithic, only to slowly decrease in frequency over time. If there is one thing we know about evolution, it is that change is eternal, so an adaptation that is useful at one time may later prove a costly burden.
Then, whole selection events were detected with clear physical implications but for which an evolutionary rationale is not immediately clear. One of the clearest patterns researchers have found in ancient DNA, and in modern genomes, is that many populations, and in particular Europeans, have been selected for lighter pigmentation in the last 10,000 years. The new findings make clear that this process has been constant over the millennia, rather than a single bout that eventually abated. Europeans were far paler in 2000 BC than in 6000 BC, and they seem to be somewhat paler today than they were 4,000 years ago. This holds true across many genetic loci implicated in pigmentation, so it cannot simply be a side effect of a separate adaptation that was actually the true selection target. The candidate motives for the selective pressure are synthesis of vitamin D, which lighter pigmentation eases, and sexual selection for paler features.
Finally, the group tackled polygenic characteristics like height, intelligence and disease risks, which all vary along smooth distributions rather than as black and white phenotypes. These outputs fluctuate due to small variations on hundreds and even thousands of separate positions in the genome. Genome-wide data has allowed scientists to understand and predict them in modern populations. The same methods from complex-trait genomics can be applied to ancient DNA. Because of the high quality of their vast dataset they were able to obtain results for many characteristics whose distributions seem to have changed over time due to selection. Predicted body-fat percentage, waist circumference, risk of bipolar disorder and schizophrenia have all decreased in Europe over the last 10,000 years. Genes associated with faster walking pace increased. Height declined with the arrival of Neolithic farmers, and rose again after the influx of Yamnaya steppe herders.
Adaptive stories do not always present themselves for these changes. Why do the genetic variants for intelligence increase sharply with the arrival of Neolithic farmers (the green point above), then decline slightly upon Yamnaya arrival (blue point), before gradually, slightly increasing up to the present? Strangely, genes associated with household income and more years of schooling in modern populations also increased. This is probably simply due to genes predicting intelligence also predicting greater income and education in modern societies (income and IQ show a 0.5 correlation). Complex traits are by their nature… complex. Even in modern populations they are not fully elucidated or understood, because they often depend on environmental interactions. Nevertheless, natural selection does not discriminate between a trait controlled by a single gene or 1,000 genes; all it sees is the variation in fitness of heritable traits. The kind of trait that continuously varies along a spectrum and gradually alters by generation is indeed the type Darwin first envisaged being shaped by natural selection, gradually and imperceptibly reshaping species.
Over the past 15 years, ancient DNA has already transformed our understanding of phylogenetics and the movements of human populations around the planet. But methods to detect natural selection require larger sample sizes and higher quality data. This preprint, and others of its kind, herald a promising new chapter in ancient DNA. 14,000 years in Europe represents a truly minuscule slice of the human adaptive story since our species’ emergence 200,000 years ago. Within the next decade, we can expect to pull off similar analyses of humanity’s tenure on the planet, from the time our species arose and across every continent it colonized, thus mapping entire global patterns of natural selection. As we advance, we can truly expect to understand human adaptation over the long arc of our history. And in so doing, we will bring to fruition the project Darwin first envisioned 150 years ago in The Descent of Man, and Selection in Relation to Sex.





Razib, a request from someone with great interest but not a background in biology/genetics.
It was wonderful to read an article of this length. Is it possible for you to write more like this in the future?